Development of 2D-QSAR Models and Molecular Docking of Protein Tyrosine Phosphatase 1B Inhibitors
Main Article Content
Abstract
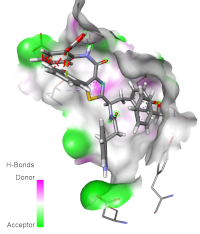
Quantitative structure-activity relationships (QSARs) are widely used in drug discovery and design to quantitatively analyze the relationships between the structures and biological activities of compounds. The present study aimed to develop a two-dimensional (2D) QSAR and molecular docking studies to predict the biological activities of protein tyrosine phosphatase 1B (PTP1B) inhibitors. A QSAR was carried out to study a series of fifty-three (53) compounds based on protein tyrosine phosphatase 1B (PTP1B) inhibitors. The study was performed using principal components analysis (PCA), multiple linear regression (MLR) and multiple non-linear regression (MNLR) to predict unambiguous QSAR models of studied compounds toward PTP1B inhibitory activity. Molecular docking was used to elucidate the inhibitory mechanisms of the most active compound from the data set against PTP1B. The statistical results of the MLR and MNLR indicate that the determination coefficients (R2) were similar (R2 = 0.796). To validate the predictive power of the resulting models, the determination coefficients of external validation were 0.815 and 0.639 for the MLR and MNLR, respectively. These results showed that both models possess favorable estimation stability and good prediction power. The docking of the most active compound 46 showed many hydrogen bond formations with the active site residues ASP (A:48) and TYR (A:46) of PTP1B. The study successfully developed a simple, convenient quantitative structure-activity relationship (QSAR) model that can be used to screen chemical databases or design new PTP1B inhibitor-derived compounds.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
1.DeFronzo RA, Bonadonna RC, Ferrannini E. Pathogenesis of NIDDM. A Balanced Overview. Diabetes Care. 1992; 15(3):318-368. doi: 10.2337/diacare.15.3.318.
2.Zimmet P, Alberti KG, Shaw J. Global and Societal Implications of the Diabetes Epidemic. Nature. 2001; 414(6865):782-787. doi: 10.1038/414782a.
3.Saltiel AR and Kahn CR. Insulin Signalling and the Regulation of Glucose and Lipid Metabolism. Nature. 2001; 414(6865):799-806. doi: 10.1038/414799a.
4.Obici S, Feng Z, Karkanias G, Baskin DG, Rossetti L. Decreasing Hypothalamic Insulin Receptors Causes Hyperphagia and Insulin Resistance in Rats. Nat Neurosci. 2002; 5(6):566-572. doi: 10.1038/nn0602-861.
5.Johnson TO, Ermolieff J, Jirousek MR. Protein Tyrosine Phosphatase 1B Inhibitors for Diabetes. Nat Rev Drug Discov. 2002; 1(9):696-709. doi: 10.1038/nrd895.
6.Drake PG and Posner BI. Insulin Receptor-Associated Protein Tyrosine Phosphatase(s): Role in Insulin Action. Mol Cell Biochem. 1998; 182(1-2):79-89.
7.White MF and Kahn CR. The Insulin Signaling System. J Biol Chem. 1994; 269(1):1-4.
8.Byon JC, Kusari AB, Kusari J. Protein-Tyrosine Phosphatase-1B Acts as a Negative Regulator of Insulin Signal Transduction. Mol Cell Biochem. 1998; 182(1-2):101-108.
9.Dadke S and Chernoff J. Interaction of Protein Tyrosine Phosphatase (PTP) 1B With Its Substrates is Influenced by Two Distinct Binding Domains. Biochem J. 2002; 364(Pt 2):377-383. doi: 10.1042/BJ20011372.
10.Bandyopadhyay D, Kusari A, Kenner KA, Liu F, Chernoff J, Gustafson TA, Kusari J. Protein-Tyrosine Phosphatase 1B Complexes with the Insulin Receptor in Vivo and is Tyrosine-Phosphorylated in the Presence of Insulin. J Biol Chem. 1997; 272(3):1639-1645. doi: 10.1074/jbc.272.3.1639.
11.Kusari J, Kenner KA, Suh KI, Hill DE, Henry RR. Skeletal Muscle Protein Tyrosine Phosphatase Activity and Tyrosine Phosphatase 1B Protein Content are Associated with Insulin Action and Resistance. J Clin Invest. 1994; 93(3):1156-1162. doi: 10.1172/JCI117068.
12.González-Díaz H. Computational Prediction of Drug-Target Interactions in Medicinal Chemistry. Curr Top Med Chem. 2013; 13(14):1619-1621. doi: 10.2174/15680266113139990112.
13.González-Díaz H, Arrasate S, Sotomayor N, Lete E, Munteanu CR, Pazos A, Besada-Porto J, Ruso JM. MIANN Models in Medicinal, Physical and Organic Chemistry. Curr Top Med Chem. 2013; 13(5):619-641. doi: 10.2174/1568026611313050006.
14.Abeijon P, Garcia-Mera X, Caamano O, Yanez M, Lopez-Castro E, Romero-Duran FJ, Gonzalez-Diaz H. Multi-Target Mining of Alzheimer Disease Proteome with Hansch's QSBR-Perturbation Theory and Experimental-Theoretic Study of New Thiophene Isosters of Rasagiline. Curr Drug Targets. 2017; 18(5):511-521. doi: 10.2174/1389450116666151102095243.
15.Todeschini R, Pazos A, Arrasate S, González-Díaz H. Data Analysis in Chemistry and Bio-Medical Sciences. Int J Mol Sci. 2016; 17(12):2105. doi: 10.3390/ijms17122105.
16.González-Díaz H, Herrera-Ibatá DM, Duardo-Sánchez A, Munteanu CR, Orbegozo-Medina RA, Pazos A. ANN Multiscale Model of Anti-HIV Drugs Activity vs AIDS Prevalence in the US at County Level Based on Information Indices of Molecular Graphs and Social Networks. J Chem Inf Model. 2014; 54(3):744-755. doi: 10.1021/ci400716y.
17.Duardo-Sánchez A, Munteanu CR, Riera-Fernández P, López-Díaz A, Pazos A, González-Díaz H. Modeling Complex Metabolic Reactions, Ecological Systems, and Financial and Legal Networks with MIANN Models Based on Markov-Wiener Node Descriptors. J Chem Inf Model. 2014; 54(1):16-29. doi: 10.1021/ci400280n.
18.Agarwal N, Bajpai A, Gupta SP. A Quantitative Structure-Activity Relationship and Molecular Modeling Study on a Series of Heteroaryl- and Heterocyclyl-Substituted Imidazo[1,2-a] Pyridine Derivatives Acting as Acid Pump Antagonists. Biochem Res Int. 2013; 2013:141469. doi: 10.1155/2013/141469.
19.Moretto AF, Kirincich SJ, Xu WX, Smith MJ, Wan ZK, Wilson DP, Follows BC, Binnun E, Joseph-McCarthy D, Foreman K, Erbe DV, Zhang YL, Tam SK, Tam SY, Lee J. Bicyclic and Tricyclic Thiophenes as Protein Tyrosine Phosphatase 1B Inhibitors. Bioorg Med Chem. 2006; 14(7):2162-2177. doi: 10.1016/j.bmc.2005.11.005.
20.Erbe DV, Klaman LD, Wilson DP, Wan ZK, Kirincich SJ, Will S, Xu X, Kung L, Wang S, Tam S, Lee J, Tobin JF. Prodrug Delivery of Novel PTP1B Inhibitors to Enhance Insulin Signalling. Diabetes Obes Metab. 2009; 11(6):579-588. doi: 10.1111/j.1463-1326.2008.01022.x.
21.Wan ZK, Lee J, Xu W, Erbe DV, Joseph-McCarthy D, Follows BC, Zhang YL. Monocyclic Thiophenes as Protein Tyrosine Phosphatase 1B Inhibitors: Capturing Interactions with Asp48. Bioorg Med Chem Lett. 2006; 16(18):4941-4945. doi: 10.1016/j.bmcl.2006.06.051.
22.Wang F and Zhou B. Toward the Identification of a Reliable 3D-QSAR Model for the Protein Tyrosine Phosphatase 1B Inhibitors. J Mol Struct. 2018; 1158:75-87. doi.org/10.1016/j.molstruc.2018.01.011.
23.Advanced Chemistry Development Inc. Toronto, Canada; 2009. www.acdlabs.com/resources/freeware/chemsketch/.
24.ChemBioOffice, PerkinElmer Informatics, 2010. http://www.cambridgesoft.com.
25.Larif M, Adad, Hmammouchi R, Taghki AI, Soulaymani, Elmidaoui A, Bouachrine M, Lakhlifi T. Biological Activities of Triazine Derivatives Combining DFT and QSAR Results. Arab J Chem. 2017; 10:s946-s955.
26.XLSTAT Software, XLSTAT Company, 2014. http://www.xlstat.com.
27.Ghamali M, Chtita S, Aouidate A, Ghaleb A, Bouachrine M, Lakhlifi T. Combining DFT and QSAR Computation to Predict the Interaction of Flavonoids with the GABA (A) Receptor Using Electronic and Topological Descriptors. J Taibah Univ Sci. 2017; 11:422-433.
28.Golbraikh A and Tropsha A. Beware of q2!. J Mol Graph Model. 2002; 20(4):269-276. doi: 10.1016/s1093-3263(01)00123-1.
29.Roy PP and Roy K. On Some Aspects of Variable Selection for Partial Least Squares Regression Models. QSAR Comb Sci. 2008; 27:302-313. doi.org/10.1002/qsar.200710043.
30.Gramatica P. Principles of QSAR Models Validation: Internal & External. QSAR Comb Sci. 2007; 26:694-701. doi.org/10.1002/qsar.200610151.
31.Jain AN. Surflex: Fully Automatic Flexible Molecular Docking Using a Molecular Similarity-Based Search Engine. J Med Chem. 2003; 46:499-511.
32.M Asmae, T Hamid, A Toufik, L Fatima. In Silico Screening of New Derivatives as Inhibitors of Enoyl-[Acyl-Carrier-Protein] Reductase From Staphylococcus Aureus Via 2D-QSAR Analysis, Molecular Docking and ADME/Tox Prediction. Trop. J. Nat. Prod. Res. 2025; 9(4):1616 – 1624.
33.Wang J, Kollman PA, Kuntz ID. Flexible Ligand Docking: A Multistep Strategy Approach. Proteins. 1999; 36(1):1-19.
34.Purcell WP and Singer JA. A Brief Review and Table of Semiempirical Parameters Used in the Hückel Molecular Orbital Method. J Chem Eng Data. 1967; 12(2):235–246. doi.org/10.1021/je60033a020.
35.Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016. http://accelrys.com/products/collaborative-science/biovia-discovery-studio/.
36.STATITCF Software, Technical Institute of Cereals and Fodder, Paris, France, 1987.
37.Erazua EA, Akintelu SA, Adelowo JM, Odoemene SN, Josiah OM, Raheem SF, Latona DF, Adeoye MD, Esan AO, Oyebamiji AK. QSAR and Molecular Docking Studies on Nitro (Triazole/Imidazole)-Based Compounds as Anti-Tubercular Agents. Trop. J. Nat. Prod. Res. 2021; 5(11):2022-2029. Doi.org/10.26538/tjnpr/v5i11.22.
38.O’Brien RM. A Caution Regarding Rules of Thumb for Variance Inflation Factors. Qual Quant. 2007; 41:673-690. doi.org/10.1007/s11135-006-901