Exploring the Protein Targets of Cinnamomum zeylanicum’s Phytoconstituents Against Pathogenic Staphylococcus aureus: GC-MS Profiling, Molecular Docking, Pharmacophore Modeling, and Pathway Analysis
DOI:
https://doi.org/10.26538/tjnpr/v9i10.48Keywords:
Pathway analysis, Molecular docking, Gas Chromatography Mass Spectrometer, Staphylococcus aureus, Cinnamomum zeylanicumAbstract
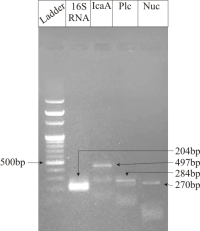
Staphylococcus aureus is a major cause of severe infections, including sepsis, largely due to its diverse virulence factors and increasing antibiotic resistance, which highlights the need for alternative therapeutic strategies. The traditional medicinal plant, Cinnamomum zeylanicum, has rich bioactive secondary metabolites. In this study, we investigated the antimicrobial potential of C. zeylanicum bark against pathogenic S. aureus through minimum inhibitory and bactericidal concentration assays, along with phytoconstituent profiling using GC-MS. Virulence factors of S. aureus were characterized by PCR targeting the plc, icaA, and nuc genes. The identified phytoconstituents were further analyzed in silico, including molecular docking, pharmacophore modeling, and ADMET analysis against S. aureus target proteins. A tetrapartite interaction network and pathway analysis were constructed using STRING and KEGG databases. The methanol extract, containing 19 phytoconstituents, exhibited significant antibacterial activity with MIC and MBC values of 5 mg/mL. Docking results revealed that α-copaene, α-muurolene, and τ-cadinol showed strong binding interactions with D-alanine-D-alanine ligase, dihydrofolate reductase, peptide deformylase, and penicillin-binding protein 2a. These findings suggest that C. zeylanicum bark extract, enriched with phenolic and flavonoid derivatives, holds promise as a natural source of anti-S. aureus agents. However, further experimental validation is needed to confirm the predicted protein targets and pathways.
References
1.Kumar, N.R., Balraj, T.A., Kempegowda, S.N., and Prashant, A. Multidrug-Resistant Sepsis: A Critical Healthcare Challenge. Antibiotics. 2024;13(1):46. doi: 10.3390/antibiotics13010046
2.Patel, H., and Rawat, S. A genetic regulatory see-saw of biofilm and virulence in MRSA pathogenesis. Front Microbiol. 2023;14:1204428. doi: 10.3389/fmicb.2023.1204428
3.Rai, A., and Khairnar, K. Overview of the risks of Staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products. Brazilian J Microbiol. 2021;52(4):2031–2042. doi: 10.1007/s42770-021-00566-4
4.Haindongo, E.H., Ndakolo, D., Hedimbi, M., Vainio, O., Hakanen, A., and Vuopio, J. Antimicrobial resistance prevalence of Escherichia coli and Staphylococcus aureus amongst bacteremic patients in Africa: a systematic review. J of Glob Antimicrob Resist. 2023;32:35–43. doi: 10.1016/j.jgar.2022.11.016
5.Eltwisy, H.O., Twisy, H.O., Hafez, M.H.R., Sayed, I.M., and El-Mokhtar, M.A. Clinical Infections, Antibiotic Resistance, and Pathogenesis of Staphylococcus haemolyticus. Microorganisms. 2022;10:1130. doi: 10.3390/microorganisms10061130
6.Zhu, Z., Hu, Z., Li, S., Fang, R., Ono, H.K., and Hu, D.L. Molecular Characteristics and Pathogenicity of Staphylococcus aureus Exotoxins. Int J Mol Sci. 2024;25(1):395. doi: 10.3390/ijms25010395
7.Yu, J., Jiang, F., Zhang, F., Hamushan, M., Du, J., Mao, Y., Wang, Q., and Shen, H. Thermonucleases contribute to Staphylococcus aureus biofilm formation in implant-associated infections–A redundant and complementary story. Front Microbiol. 2021;12:687888. doi: 10.3389/fmicb.2021.687888
8.Yu, J., Han, W., Xu, Y., Shen, L., Zhao, H., Zhang, J., Xiao, Y., Guo, Y., and Yu, F. Biofilm-producing ability of methicillin-resistant Staphylococcus aureus clinically isolated in China. BMC Microbiol. 2024;24(1):241. doi: 10.1186/s12866-024-03380-8
9.Singh, I., Roshan, M., Vats, A., Behera, M., Gautam, D., Rajput, S., Rana, C., and De, S. Evaluation of Virulence, Antimicrobial Resistance and Biofilm Forming Potential of Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Bovine Suspected with Mastitis. Curr Microbiol. 2023;80(6):198. doi: 10.1007/s00284-023-03303-2
10.Hou, Z., Liu, L., Wei, J., and Xu, B. Progress in the Prevalence, Classification and Drug Resistance Mechanisms of Methicillin-Resistant Staphylococcus aureus. Infect Drug Resist. 2023;16:3271–3292. doi: 10.2147/IDR.S412308
11.Gu, D.T., Tung, T.H., Jiesisibieke, Z.L., Chien, C.W., and Liu, W.Y. Safety of Cinnamon: An Umbrella Review of Meta-Analyses and Systematic Reviews of Randomized Clinical Trials. Front Pharmacol. 2022;12(18):790901. doi: 10.3389/fphar.2021.790901
12.Ribeiro-santos, R., Andrade, M., Madella, D., Melo, D., Paula, A., Aquino, L. De, Moura, G., and Sanches-silva, A. Revisiting an ancient spice with medicinal purposes: Cinnamon. Trends Food Sci Technol. 2017;62:154–169. doi: 10.1016/j.tifs.2017.02.011
13.Khalisyaseen, O., and Mohammed, M.T. Analytics Detection of Phytochemical Compounds in Cinnamomum zeylanicum Bark Extract. Egypt J Chem. 2023;66(4):265–273. doi: 10.21608/EJCHEM.2022.146416.6366
14.Prabhashini, W., Mendis, K., Arachchige, G., and Premakumara, S. Anti‑inflammatory, cytotoxicity and antilipidemic properties : novel bioactivities of true cinnamon (Cinnamomum zeylanicum Blume) leaf. BMC Complement Med Ther. 2022;22:259. doi: 10.1186/s12906-022-03728-5
15.Alizadeh Behbahani, B., Falah, F., Lavi Arab, F., Vasiee, M., and Tabatabaee Yazdi, F. Chemical Composition and Antioxidant, Antimicrobial, and Antiproliferative Activities of Cinnamomum zeylanicum Bark Essential Oil. J Evid Based Complement Altern Med. 2020;2020:1590603. doi: 10.1155/2020/5190603
16.Nawaz, A., Ali, T., Naeem, M., Hussain, F., Li, Z., and Nasir, A. Biochemical, structural characterization and in-vitro evaluation of antioxidant, antibacterial, cytotoxic, and antidiabetic activities of nanosuspensions of Cinnamomum zeylanicum bark extract. Front Chem. 2023;11:1194389. doi: 10.3389/fchem.2023.1194389
17.Quyen, P.T., and Quoc, L.P.T. Chemical Profile and Biological Activities of The Essential Oil of Cinnamon (Cinnamomum cassia (L.) J. Presl) Twigs and Leaves. Trop J Nat Prod Res. 2023;7(11):5226–5230. doi: 10.26538/tjnpr/v7i11.29
18.Rollando, R., Eva, M., Dodi, I., Viol, D.K., and Arif, N.M. Goniothalamus macrophyllus: A Comprehensive Review of Its Phytochemistry, Pharmacological Activities, and Therapeutic Potential. Trop J Nat Prod Res. 2025;7(9):2964–2972. doi: 10.26538/tjnpr/v9i7.3
19.Mulpuru, V., and Mishra, N. Computational Identification of SARS-CoV-2 Inhibitor in Tinospora cordifolia, Cinnamomum zeylanicum and Myristica fragrans. VirusDisease. 2021;32(3):511–517. doi: 10.1007/s13337-021-00721-3
20.Meylani, V., Rizal Putra, R., Miftahussurur, M., Sukardiman, S., Eko Hermanto, F., and Abdullah, A. Molecular docking analysis of Cinnamomum zeylanicum phytochemicals against Secreted Aspartyl Proteinase 4–6 of Candida albicans as anti-candidiasis oral. Results Chem. 2023;5:100721. doi: 10.1016/j.rechem.2022.100721
21.Pourkhosravani, E., Nayeri, F.D., and Bazargani, M.M. Decoding antibacterial and antibiofilm properties of cinnamon and cardamom essential oils : a combined molecular docking and experimental study. AMB Express. 2021;11:143. doi: 10.1186/s13568-021-01305-6
22.Julianti, E., Rajah, K.K., and Fidrianny, I. Antibacterial activity of ethanolic extract of Cinnamon bark, honey, and their combination effects against acne-causing bacteria. Sci Pharm. 2017;85(2):19. doi: 10.3390/scipharm85020019
23.Mujeeb, F., Bajpai, P., and Pathak, N. Phytochemical Evaluation, Antimicrobial Activity, and Determination of Bioactive Components from Leaves of Aegle marmelos. BioMed Res Int. 2014;2014:497606. doi: 10.1155/2014/497606
24.Dvorackova, E., Snoblova, M., Chromcova, L., and Hrdlicka, P. Effects of extraction methods on the phenolic compounds contents and antioxidant capacities of cinnamon extracts. Food Sci Biotechnol. 2015;24(4):1201–1207. doi: 10.1007/s10068-015-0154-4
25.Rugaie, O. Al, Mohammed, H.A., Alsamani, S., Messaoudi, S., Aroua, L.M., Khan, R.A., Almahmoud, S.A., and Altaleb, A.D. Antimicrobial , Antibiofilm , and Antioxidant Potentials of Four Qassim Region of Saudi Arabia : Phytochemical Profile and In Vitro and In Silico Bioactivity Investigations. Antibiotics. 2023;12(3):501. doi: 10.3390/antibiotics12030501
26.Rhetso, T., Seshadri, R.M., Ramnath, S., and Venkataramegowda, S. GC-MS based metabolite profiling and antioxidant activity of solvent extracts of Allium Chinense G Don leaves. Not Sci Biol. 2021;13(2):10791. doi: 10.15835/nsb13210791
27. Precious, A.I., Joan, M.O., David, A.Z., Fatima, A.S., Ummulkhairi, T., Endaline, A.M., Obumneme. C.O., Abubakar, R.M., and Samson, C.O. Antidiabetic Activity and in silico Molecular Docking of GC-MS-Identified Compounds in Chromatographic Fractions of Tephrosia bracteolata Guill. & Perr.(Fabaceae) Leaves. Trop J Nat Prod Res. 2025;9(8):3720–3728. doi: 10.26538/tjnpr/v9i8.31
28.Brakstad, O.D.D.G., Aasbakk, K., and Maeland, J.A. Detection of Staphylococcus aureus_nuc pcr und primer. J Clin microbiol. 1992;30(7):1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992
29.Mith, H., Duré, R., Delcenserie, V., Zhiri, A., Daube, G., and Clinquart, A. Antimicrobial activities of commercial essential oils and their components against food‐borne pathogens and food spoilage bacteria. Food Sci Nutr. 2014;2(4):403–416. doi: 10.1002/fsn3.116
30.Prabha, S., Chauhan, P., Warkare, S., and Pandey, K.M. A computational investigation of potential plant-based bioactive compounds against drug-resistant Staphylococcus aureus of multiple target proteins. J Biomol Struct Dyn. 2023;43(7):3311–3329. doi: 10.1080/07391102.2023.2297009
31.Kirchmair, J., Markt, P., Distinto, S., Schuster, D., Spitzer, G.M., Liedl, K.R., Langer, T., and Wolber, G. The Protein Data Bank (PDB), Its Related Services and Software Tools as Key Components for In Silico Guided Drug Discovery. J Med Chem. 2008;51(22):7021–7040. doi: 10.1021/jm8005977
32.Trott, O., and Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31(1):455–461. doi: 10.1002/jcc.21334
33.Csizmadia, P. MarvinSketch and MarvinView: Molecule Applets for the World Wide Web, in Proceedings of the 3rd International Electronic Conference on Synthetic Organic Chemistry. MDPI:Basel, Swit. 1999. doi: 10.3390/ecsoc-3-01775
34.Boyle, N.M.O., Banck, M., James, C.A., Morley, C., Vandermeersch, T., and Hutchison, G.R. Open Babel : An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33
35.Maćkiewicz, A., and Ratajczak, W. Principal components analysis (PCA). Computers & Geosciences. 1993;19(3):303–342. doi: 10.1016/0098-3004(93)90090-R
36.Alin, A. Minitab. WIREs Comput Stat. 2010;2(6):723–727. doi: 10.1002/wics.113
37.Kohl, M., Wiese, S., and Warscheid, B. Cytoscape: Software for Visualization and Analysis of Biological Networks. In: Hamacher, M., Eisenacher, M., Stephan, C. (eds) Data Mining in Proteomics. Methods Mol Bio. 2011;696. doi: 10.1007/978-1-60761-987-1_18
38.Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth, A., Minguez, P., Doerks, T., Stark, M., Muller, J., Bork, P., Jensen, L.J., and Mering, C. von. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucl Acids Res. 2010;39(suppl_1):D561–D568. doi: 10.1093/nar/gkq973
39.Aoki, K.F., and Kanehisa, M. Using the KEGG Database Resource. Current Protocols in Bioinforma. 2005;11:1.12.1-1.12.54. doi: 10.1002/0471250953.bi0112s11
40.Daina, A., Michielin, O., and Zoete, V. SwissADME : a free web tool to evaluate pharmacokinetics , drug- likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717
41.Banerjee, P., Eckert, A.O., Schrey, A.K., and Preissner, R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucl Acids Res. 2018;46(W1):W257–W263. doi: 10.1093/nar/gky318
42.Lang, G., and Buchbauer, G. A review on recent research results (2008–2010) on essential oils as antimicrobials and antifungals. A review. Flavour Fragr J. 2012;27(1):13–39. doi: 10.1002/ffj.2082
43.43. Ranjan Sahoo, C., Sahoo, J., Mahapatra, M., Lenka, D., Kumar Sahu, P., Dehury, B., Nath Padhy, R., and Kumar Paidesetty, S. Coumarin derivatives as promising
antibacterial agent(s). Arab J Chem. 2021;14(2):102922. doi: 10.1016/j.arabjc.2020.102922
44.Santos, A.L. dos, Amaral, M., Hasegawa, F.R., Lago, J.H.G., Tempone, A.G., and Sartorelli, P. (-)-T-Cadinol—a Sesquiterpene Isolated From Casearia sylvestris (Salicaceae)—Displayed In Vitro Activity and Causes Hyperpolarization of the Membrane Potential of Trypanosoma cruzi. Front Pharmacol. 2021;12:734127. doi: 10.3389/fphar.2021.734127
45.Komane, B., Kamatou, G., Mulaudzi, N., Vermaak, I., and Fouche, G. Chapter 21 - Sclerocarya birrea. The south Afri herb Pharmacopoeia. 2023:471-501. doi: 10.1016/B978-0-323-99794-2.00027-1
46.Abdel-Shafi, S., El-Serwy, H., El-Zawahry, Y., Zaki, M., Sitohy, B., and Sitohy, M. The Association between icaA and icaB Genes, Antibiotic Resistance and Biofilm Formation in Clinical Isolates of Staphylococci spp. Antibiotics. 2022;11(3):389. doi: 10.3390/antibiotics11030389
47.Nakamura, Y., Kanemaru, K., Shoji, M., Totoki, K., Nakamura, K., Nakaminami, H, Nakase K, Noguchi N & Fukami K. Phosphatidylinositol-specific phospholipase C enhances epidermal penetration by Staphylococcus aureus. Sci Rep. 2020;10:17845. doi: 10.1038/s41598-020-74692-8
48.Kiedrowski, M.R., Crosby, H.A., Hernandez, F.J., Malone, C.L., McNamara, J.O., and Horswill, A.R. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS ONE. 2014;9(4):e95574. doi: 10.1371/journal.pone.0095574
49.Devara, M.S., Bhamidipati, S., Dondapati, V.B., and Bandaru, N.R. Antibacterial activity of cinnamon extract against gram-positive and gram-negative bacterial pathogens isolated from patient samples. Med. Lab. J. 2023;17(6):1–3. doi: 10.61186/mlj.17.6.1
50.Tamiru T, D.T., and Belete D, B.B. Antimicrobial Potentials of Apis Multiflora Honey in Combination with Coffee and Cinnamon Extracts against Common Human Pathogenic Bacteria. Med. Aromat. Plant. 2015;4(4):208. doi: 10.4172/2167-0412.1000208
51.Shaker, B., Yu, M.S., Song, J.S., Ahn, S., Ryu, J.Y., Oh, K.S., Na, Dokyun. LightBBB: Computational prediction model of blood-brain-barrier penetration based on LightGBM. Bioinformatics. 2021;37(8):1135–1139. doi: 10.1093/bioinformatics/btaa918
Published
Issue
Section
License
Copyright (c) 2025 Tropical Journal of Natural Product Research

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.







