Enhancement of the Antibiofilm Activity Against Pseudomonas aeruginosa and Biodiesel Production Using Lipase from Cronobacter dublinensis as a Catalyst
Main Article Content
Abstract
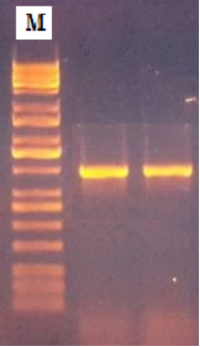
Lipases can convert edible and vegetable oils into biodiesel, a low-toxicity, renewable, biodegradable, and environmentally friendly alternative to traditional fuel. This study investigated the dual functionality of lipase from Cronobacter dublinensis in enhancing antibiofilm activity against Pseudomonas aeruginosa and catalyzing biodiesel production via enzymatic transesterification. Cronobacter dublinensis was isolated from lettuce samples and identified by morphological, biochemical, and molecular methods, with gyrB gene confirmation. Lipase production was screened on oil-supplemented agar, and the gene known as Lip was detected via polymerase chain reaction (PCR). The lipase was then purified using ammonium sulfate precipitation in combination with dialysis and chromatographic techniques. Antibiofilm activity was assessed using a microtiter plate method against Pseudomonas aeruginosa. Biodiesel production was performed via enzymatic transesterification using crude lipase. Four Cronobacter dublinensis isolates were identified from the lettuce samples. Lipase production induced by various oils revealed that olive oil yielded the highest hydrolytic activity, producing zones of 20–50 mm. The Lip gene was detected in all the isolates via PCR (550 bp fragment). Purification through the combined use of ammonium sulfate precipitation and chromatography resulted in a 5.5-times improvement in purity with 58% recovery. Purified lipase significantly inhibited the establishment of biofilms by Pseudomonas aeruginosa, which varied with dosage, showing maximum suppression at a concentration of 200 μg/mL. Enzymatic transesterification using C. dublinensis lipase achieved 38% and 11.2% fatty acid methyl ester yields for processed olive and corn oils, respectively. These findings highlight the dual potential of C. dublinensis lipase in antimicrobial and biofuel applications.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
1. Ali S, Khan SA, Hamayun M, Lee IJ. The recent advances in the utility of microbial lipases: A review. Microorganisms. 2023; 11(2):510-523.
2. Hu Z, Jiao L, Xie X, Xu L, Yan J, Yang M, Yan Y. Characterization of a new thermostable and organic solution-tolerant lipase from Pseudomonas fluorescens and its application in the enrichment of polyunsaturated fatty acids. Int J Mol Sci. 2023; 24(10):8924-8931.
3. Muslim SN. Purification and partial characterization of extracellular lipase from Pseudomonas fluorescens. Al-Mustansiriya J Sci. 2007; 18(3):46–61.
4. Muslem WH, Muslim SN, Mahammed Ali AN, Fayyad RJ. Detection of disinfectant property of purified amylopullulanase from Citrobacter freundii SW. Research J. Pharm. and Tech. 2022; 15(2): 847-852.
5. Fukuda H, Kondo A, Noda H. Biodiesel fuel production by transesterification of oils. J Biosci Bioeng. 2001; 92:405–416.
6. Ghaly AE, Dave D, Brooks MS, Budge S. Production of biodiesel by enzymatic transesterification: Review. Am J Biochem Biotechnol. 2010; 6(2):54–76.
7. Orege JI, Oderinde O, Kifle GA, Ibikunle AA, Raheem SA, Ejeromedoghene O, Okeke ES, Olukowi OM, Orege OB, Fagbohun EO, Ogundipe TO, Praise Avor EP, Ajayi OO, Daramola MO. Recent advances in heterogeneous catalysis for green biodiesel production by transesterification. Energy Convers Manag. 2022; 258(115406):1-11.
8. El-Hadi AA, Saleh HI, Moustafa SA, Ahmed HM. Immobilization of Mucor racemosus NRRL 3631 lipase and characterization of silica-coated magnetite (Fe₃O₄) nanoparticles. Egypt Pharm J. 2013; 12:28–35.
9. Muslim SN, Al-Kadmy IMS, Auda IG, Mohammed Ali AN, Al-Jubori SS. A novel genetic Determination of a lectin gene in Iraqi Acinetobacter baumannii isolates and use of purified lectin as an antibiofilm agent. J. AOAC Intern. 2018; 101(1):1–8.
10. Turinayo YK, Kalanzi F, Mudoma JM, Kiwuso P, Asiimwe GM, Esegu JPO, Balitta P, Mwanja C. Physicochemical characterization of Jatropha curcas Linn oil for biodiesel production in Nebbi and Mokono districts in Uganda. J Sustain Bioenergy Syst. 2015; 5:104–113.
11. Farouk SM, Tayeb AM, Abdel-Hamid SMS. Recent advances in transesterification for sustainable biodiesel production, challenges, and prospects: a comprehensive review. Environ Sci Pollut Res. 2024; 31:12722–12747.
12. Ghanbari A, Dehghany J, Schwebs T, Müsken M, Häussler S, Meyer-Hermann M. Inoculation density and nutrient level determine the formation of mushroom-shaped structures in Pseudomonas aeruginosa biofilms. Sci Rep. 2016; 6(1):1–12.
13. Muslim SN, Mohammed Ali AN, Auda IG. Anti‐biofilm and anti‐virulence effects of silica oxide nanoparticle–conjugation of lectin purified from Pseudomonas aeruginosa. IET Nanobiotechnol. 2021; 15(3):1–11.
14. Gholami S, Tabatabaei M. The prevalence of virulence factors in human and environmental isolates of Pseudomonas aeruginosa. Avicenna J Clin Microbiol Infect. 2019; 6(1):9–14.
15. Ling, N.; Li,C.; Zhang, J.; Wu, Q.; Zeng, H.; He, W.; Ye, Y.; Wang, J.; Ding, Y.; Chen, M.; Xue, L.; Ye, Q. and Guo, W.(2018). Prevalence and molecular and antimicrobial characteristics of Cronobacter spp. isolated from raw vegetables in China. Front Microbiol. 2018; 9:1-10.
16. Ruppé E, Hem S, Lath S, Gautier V, Ariey F, Sarthou JL, et al. CTX-M β-lactamases in Escherichia coli from community-acquired urinary tract infections, Cambodia. Emerg Infect Dis. 2009; 15(5):741–748.
17. Al Mohaini M, Farid A, Muzammal M, Ghazanfar S, Dadrasnia A, Alsalman AJ, et al. Enhancing lipase production of Bacillus salmalaya strain 139SI using different carbon sources and surfactants. Appl Microbiol. 2022; 2:237–247.
18. Ali AA, Hameed KW, Nadder MI. Production, purification and biochemical characterization of lipase from Pseudomonas aeruginosa. Iraqi J Biotechnol. 2021; 20(1):70–77.
19. Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976; 72:248–54.
20. Krieg NR, Holt JG. Bergey’s Manual for Systematic Bacteriology, Vol. 2. Baltimore (MD): Williams and Wilkins; 2001; 721p.
21. Zhang D, Xia J, Xu Y, Gong M, Zhou Y, Xie L, Fang X. Biological features of biofilm-forming ability of Acinetobacter baumannii strains derived from 121 elderly patients with hospital-acquired pneumonia. Clin Exp Med. 2016; 16(1):73–80.
22. Ahmad F, Khan AU, Yasar A. Transesterification of oil extracted from different species of algae for biodiesel production. Afr J Environ Sci Technol. 2013; 7(6):358–364.
23. Escobar-Nin A, Luna C, Luna D, Marcos AT, Cánovas D, Mellado EO. Selection and characterization of biofuel-producing environmental bacteria isolated from vegetable oil-rich wastes. PLoS One. 2014; 9(8):1–8.
24. Kamal SM, Maharik NSM, Valero A, Faried AM. Prevalence and molecular characterization of Cronobacter species in Egyptian table eggs and egg-based desserts; special insight on Cronobacter sakazakii. Ital J Food Sci. 2023; 35(4):50–57.
25. Breeuwer P, Lardeau A, Peterz M, Joosten HM. Desiccation and heat tolerance of Enterobacter sakazakii. J Appl Microbiol. 2003; 95:967–973.
26. Belal M, Al-Mariri A, Hallab L, Hamad I. Detection of Cronobacter spp. (formerly Enterobacter sakazakii) from medicinal plants and spices in Syria. J Infect Dev Ctries. 2013; 7(2):82–89.
27. Nunes PM, Martinsa AB, Brígida AI S, Miguez da Rocha-Leãoa MH, Amaral P. Intracellular lipase production by Yarrowia lipolytica using different carbon sources. Chem Eng Trans. 2018; 38:421–426.
28. Paludo MP, Degani de Oliveira KS, Trevisol TC, Fernandes de Medeiros Burkert J. Isolation of lipase-producing yeasts from industrial oily residues in different culture media. Int J Curr Microbiol Appl Sci. 2018; 7:2348–2362.
29. Sipiczki G, Micevic SS, Kohari-Farkas C, Nagy ES, Nguyen QD, Gere A, Bujna E. Effects of olive oil and Tween 80 on production of lipase by Yarrowia yeast strains. Processes. 2024; 12:1206-1211.
30. Liu W, Li M, Yan Y. Heterologous expression and characterization of a new lipase from Pseudomonas fluorescens Pf0-1 and used for biodiesel production. Sci Rep. 2017; 7(1):1–11.
31. Abdou AM. Purification and partial characterization of Serratia marcescens lipase. J Dairy Sci. 2003; 86:127–132.
32. Fazeli H, Akbari R, Moghim S, Narimani T, Arabestani MR, Ghoddousi AR. Pseudomonas aeruginosa infections in patients, hospital means, and personnel's specimens. J Res Med Sci. 2021; 17(4):332–337.
33. Saha K, Kabir ND, Islam MR, Amin MB, Hoque KI, Halder K, Saleh AA, Parvez MAK, Begum K, Alam MJ, Islam MA. Isolation and characterization of carbapenem-resistant Pseudomonas aeruginosa from hospital environments in tertiary care hospitals in Dhaka, Bangladesh. J Glob Antimicrob Resist. 2022; 30:31–37.
34. Ezeador CO, Ejikeugwu PC, Ushie SN, Agbakoba NR. Isolation, identification and prevalence of Pseudomonas aeruginosa isolates from clinical and environmental sources in Onitsha Metropolis, Anambra State. Eur J Med Health Sci. 2020; 2(2):29-35.
35. De Silva K, Calomino MA, Deutsch G, de Castilho SR, de Paula GR, Esper L, Teixeira LA. Molecular characterization of multidrug-resistant (MDR) Pseudomonas aeruginosa isolated in a burn center. Burns. 2017; 43(1):137–143.
36. Yekani M, Memar MY, Alizadeh N, Safaei N, Ghotaslou R. Antibiotic resistance patterns of biofilm-forming Pseudomonas aeruginosa isolates from mechanically ventilated patients. Int J Sci Study. 2017; 5(5):1–5.
37. Behzadi P, Baráth Z, Gajdács M. It’s not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics. 2021; 10(1):1-21.
38. Gerday C, Aittaleb M, Arpigny JL, Baise E, Chessa JP,
39. Garssoux G, Petrescu I, Feller G. Psychrophilic enzymes: a thermodynamic challenge. Biochim Biophys Acta. 1997; 1342:119–131.
40. Joseph B, Ramteke PW, Thomas G, Shrivastava N. Cold-active microbial lipases: a versatile tool for industrial applications. Biotechnol Mol Biol Rev. 2007;2(2):39–48.
41. Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006; 39:235– 251.
42. Liu KS. Preparation of fatty acid methyl esters for gas chromatographic analysis of lipids in biological materials. J Am Chem Soc. 1994; 71:1179–1187.
43. Narwal SK, Gupta R. Biodiesel production by transesterification using immobilized lipase. Biotechnol Lett. 2019; 35(4):479–490.
44. Akinsemolu AA, Onyeaka HN. Microorganisms associated with food spoilage and foodborne diseases. In: Food safety and quality in the global south. Singapore: Springer Nature Singapore; 2024; pp. 489–531.
45. Sipiczki G, Micevic SS, Kohari-Farkas C, Nagy ES, Nguyen QD, Gere A, Bujna E. Effects of olive oil and Tween 80 on production of lipase by Yarrowia yeast strains. Processes. 2024; 12(6):1206-1214.
46. Mobarak-Qamsari E, Kasra-Kermanshahi R, Moosavi-Nejad Z. Isolation and identification of a novel, lipase-producing bacterium, Pseudomonas aeruginosa KM110. Iran J Microbiol. 2011; 3(2):92–99.
47. García-Silvera EE, Martínez-Morales F, Bertrand B, Morales-Guzmán D, Rosas-Galván NS, León-Rodríguez R, Trejo-Hernández MR. Production and application of a thermostable lipase from Serratia marcescens in detergent formulation and biodiesel production. Biotechnol Appl Biochem. 2018; 65(2):156–172.
48. Al-Madboly LA, Aboulmagd A, El-Salam MA, Kushkevych I, El-Morsi RM. Microbial enzymes as powerful natural anti-biofilm candidates. Microb Cell Fact. 2024; 23(1):343-349.
49. Cavali M, Bueno A, Fagundes AP, Priamo WL, Bilibio D, Mibielli GM, Perondi D, Scharf DR, Mazutti MA, Oliveira JV. Liquid lipase-mediated production of biodiesel from agroindustrial waste. Biocatal Agric Biotechnol. 2020; 30:101864.