In Silico Evaluation of Quercetin and CAPE as Potential Anti-Inflammatory Agents Targeting COX-2
DOI:
https://doi.org/10.26538/tjnpr/v9i10.4Keywords:
Propolis, Quercetin, Caffeic Acid Phenethyl Ester, Anti-inflammatory, Cyclooxygenase-2Abstract
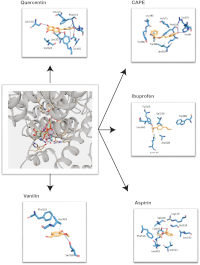
Propolis, has emerged as a promising candidate in this regard due to its active compounds, such as quercetin and caffeic acid phenethyl ester (CAPE).7,8 These compounds exhibit anti-inflammatory, making propolis a potential alternative to conventional nonsteroidal anti-inflammatory drugs (NSAID). This study evaluates the anti-inflammatory potential of quercetin and CAPE, compounds from propolis, through in silico analysis focusing on cyclooxygenase-2 (COX-2) inhibition. Toxicity predictions, molecular docking, and molecular dynamics simulations were used to assess their safety and efficacy. The COX-2 structure (PDB ID: 4PH9) was retrieved from the Protein Data Bank, and ligands (quercetin, CAPE, ibuprofen, aspirin, and vanillin) were prepared for docking. Toxicity assessments were conducted using PROTOX II and pkCSM, including AMES, LD50, skin sensitization, and hepatotoxicity tests. Molecular docking was performed using Dock 6.8, and protein-ligand interactions were validated by RMSD. Molecular dynamics simulations were carried out with Amber22, including energy minimization, solvation, and stability evaluation via RMSD, RMSF, and MM-GBSA calculations. Toxicity predictions indicated that quercetin and CAPE are safe, with negative results in mutagenicity, skin sensitization, and hepatotoxicity tests, and LD50 values supporting their therapeutic potential. Molecular docking showed strong binding of quercetin (-51.4 kcal/mol) and CAPE (-53.3 kcal/mol) to COX-2, outperforming ibuprofen and aspirin. Molecular dynamics simulations revealed stable protein-ligand interactions, with low RMSD (0.3–0.4 Å) and RMSF values, indicating high stability. MM-GBSA calculations confirmed the strong binding of quercetin (-35.4 kcal/mol) and CAPE (-33.0 kcal/mol). Quercetin and CAPE exhibit strong COX-2 binding and stability, alongside favorable safety profiles, supporting their potential as anti-inflammatory agents for further clinical investigation.
References
Jain N, Dutt U, Radenkov I, Jain S. WHO’s global oral health status report 2022: Actions, discussion and implementation. *Oral Dis.* 2024; 30(2): 73–79.
Kale S, Kakodkar P, Shetiya S, Abdulkader R. Prevalence of dental caries among children aged 5–15 years from 9 countries in the Eastern Mediterranean Region: a meta-analysis. *East Mediter Health J.* 2020; 26(6): 726–735.
Diep MT, Skudutyte-Rysstad R, Sødal ATT, Young A, Hove LH. Caries experience and risk indicators of having decayed teeth among 65-year-olds in Oslo, Norway: a cross-sectional study. *BMC Oral Health.* 2023; 23(1): 726.
Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic Diseases Caused by Oral Infection. *Clin Microbiol Rev.* 2000 Oct 1; 13(4): 547–558.
Zarghi A, Arfaei S. Selective COX-2 Inhibitors: A Review of Their Structure-Activity Relationships. *Iran J Pharm Res.* 2011; 10(4): 655–683.
Bjarnason I, Scarpignato C, Holmgren E, Olszewski M, Rainsford KD, Lanas A. Mechanisms of Damage to the Gastrointestinal Tract From Nonsteroidal Anti-Inflammatory Drugs. *Gastroenterology.* 2018; 154(3): 500–514.
Heidari G, Najafpour GD, Mohammadi M, Moghadamnia AA. Microwave Ultrasound Assisted Extraction: Determination of Quercetin for Antibacterial and Antioxidant Activities of Iranian Propolis. *Int J Engineer.* 2019; 32(8): 1057–1064.
Jing Wu MK. Propolis and its Active Component, Caffeic Acid Phenethyl Ester (CAPE), Modulate Breast Cancer Therapeutic Targets via an Epigenetically Mediated Mechanism of Action. *J Cancer Sci Ther.* 2013; 05(10): 334–342.
Chandna P, Adlakha V, Das S, Singh S. Complementary and Alternative Medicine (CAM): A Review of Propolis in Dentistry. *Am J Phytomed Clin Ther.* 2014; 2(6): 670–685.
Chang Y, Hawkins BA, Du JJ, Groundwater PW, Hibbs DE, Lai F. A Guide to In Silico Drug Design. *Pharmaceutics.* 2022; 15(1): 49.
Adelusi TI, Oyedele A-QK, Boyenle ID, Ogunlana AT, Adeyemi RO, Ukachi CD, et al. Molecular modeling in drug discovery. *Inform Med Unlocked.* 2022; 29: 100880.
De Vivo M, Masetti M, Bottegoni G, Cavalli A. Role of Molecular Dynamics and Related Methods in Drug Discovery. *J Med Chem.* 2016; 59(9): 4035–4061.
Arundina I, Frimayanti N, Surboyo MDC, Budhy TI, Iskandar B. 6-Octadecenoic and Oleic Acid in Liquid Smoke Rice Husk Showed COVID-19 Inhibitor Properties. Tlili N, editor. *Adv Pharmacol Pharm Sci.* 2024; 2024: 1–9.
Arundina I, Frimayanti N, Surboyo MDC, Budhy TI, Iskandar B, Pradana Al. In Silico Study of Liquid Smoke Rice Husk against COVID-19. *Eur J Dent.* 2023; 17(2): 492–496.
Mohammad Iqbal, R Veryanto Kurniawan, Helmi Dwi Wahyu Nurfani, Retno Indrawati Roestamadji, Muhammad Luthfi, Dini Setyowati. Molecular docking analysis of major active compounds of pomegranate peel extract (*Punica granatum* L.) in inhibiting cyclooxygenase enzyme. *World J Adv Res Rev.* 2023; 20(3): 1824–1842.
Kufareva I, Abagyan R. Methods of Protein Structure Comparison. In: *Methods in Molecular Biology.* 2011: 231–257.
Noviardi H, Fachrurrazie F. Potential of the Compound Bullatalisin as an Inhibitor of Leukotriene A4 Hydrolase Protein in Colon Cancer: An In Silico Study. *Fitofarmaka: J Ilmiah Farm.* 2015; 5(2): 65–73.
Li AP. Screening for human ADME/Tox drug properties in drug discovery. *Drug Discov Today.* 2001; 6(7): 357–366.
Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil CR. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. *Br J Pharmacol.* 2008; 153(S1).
Bell EW, Zhang Y. DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. *J Cheminform.* 2019; 11(1): 40.
Poli G, Martinelli A, Tuccinardi T. Reliability analysis and optimization of the consensus docking approach for the development of virtual screening studies. *J Enzyme Inhib Med Chem.* 2016; 31(sup2): 167–173.
Kreutzer AG, Hamza IL, Spencer RK, Nowick JS. X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an Aβ 17–36 β-Hairpin. *J Am Chem Soc.* 2016; 138(13): 4634–4642.
Az-Zahra F, Afidika J, Diamantha SDA, Rahmani AE, Fatimah S, Aulifa DL. In Silico Study of Betel Leaves Compound (*Piper betle* L.) as Acetylcholinesterase (AChE) Enzyme Inhibitor in Alzheimer Disease. *Indones J Biol Pharm.* 2022; 2(2): 44–58.
Zheng L, Meng J, Jiang K, Lan H, Wang Z, Lin M. Improving protein–ligand docking and screening accuracies by incorporating a scoring function correction term. *Brief Bioinform.* 2022; 23(3): 1–15.
Boittier ED, Tang YY, Buckley ME, Schuurs ZP, Richard DJ, Gandhi NS. Assessing Molecular Docking Tools to Guide Targeted Drug Discovery of CD38 Inhibitors. *Int J Mol Sci.* 2020; 21(15): 5183.
Limongelli V, Bonomi M, Marinelli L, Gervasio FL, Cavalli A, Novellino E. Molecular basis of cyclooxygenase enzymes (COXs) selective inhibition. *Proc Nat Acad Sci.* 2010; 107(12): 5411–5416.
Elengoe A, Naser M, Hamdan S. Modeling and Docking Studies on Novel Mutants (K71L and T204V) of the ATPase Domain of Human Heat Shock 70 kDa Protein 1. *Int J Mol Sci.* 2014; 15(4): 6797–6814.
Dermawan D, Sumirtanurdin R, Dewantisari D. Molecular Dynamics Simulation Estrogen Receptor Alpha against Andrographolide as Anti Breast Cancer. *Ind J Pharm Sci Tech.* 2019; 6(2): 65.
Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. *Expert Opin Drug Discov.* 2015; 10(5): 449–461.
Published
Issue
Section
License
Copyright (c) 2025 Tropical Journal of Natural Product Research

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.







