The Exploration of 6-Gingerol and 6-Shogaol using Network Pharmacology and Molecular Docking as Potential Inhibitors of Atherogenesis
Main Article Content
Abstract
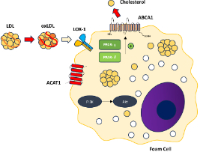
Atherosclerosis is the narrowing of blood vessels due to fatty plaque buildup in the intimal layer. Preventing atherosclerosis is a promising treatment avenue. Research shows that ginger (Zingiber officinale), particularly its active compounds 6-gingerol and 6-shogaol, can lower total cholesterol and LDL (low-density lipoprotein) levels, although the mechanisms remain unclear. The Comparative Toxigenomics Database was used to identify genes interacting with 6-gingerol and 6-shogaol, followed by enrichment analysis via ShinyGO and mapping onto KEGG pathways to identify upstream protein regulators. Docking studies were conducted using AutoDock 4.2, with visualization in Discovery Studio. Five proteins emerged as potential targets: PPAR-γ (peroxisome proliferator-activated receptor gamma), PPAR-δ (peroxisome proliferator-activated receptor delta), LOX-1 (lectin-like oxidized low-density lipoprotein receptor 1), ACAT1 (acetyl-CoA acetyltransferase 1), and PI3K (phosphoinositide 3-kinase gamma). 6-gingerol and 6-shogaol inhibited PI3K, PPAR-δ, and LOX-1 similarly to their co-crystallized ligands. However, the docking protocols could not account for LOX-1 tetramerization or ACAT1 steric hindrance, highlighting the need for further investigation into these interactions. Regarding PPAR-γ, the compounds did not show compatible interaction patterns, making it an unlikely target. The experiment provides insight into how 6-gingerol and 6-shogaol may affect lipid metabolism and atherosclerosis, mainly interacting with PI3K, PPAR-δ, and LOX-1. No supporting evidence was found for their interaction with the other tested proteins.
Downloads
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
References
1. Jamkhande PG, Chandak PG, Dhawale SC, Barde SR, Tidke PS, Sakhare RS. Therapeutic approaches to drug targets in atherosclerosis. Saudi Pharm J. 2014;22(3):179-190. doi:10.1016/j.jsps.2013.04.005
2. Jebari-Benslaiman S, Galicia-García U, Larrea-Sebal A, Olaetxea JR, Alloza I, Vandenbroeck K, Benito-Vicente A, Martín C. Pathophysiology of Atherosclerosis. Int J Mol Sci. 2022;23(6):3346. doi:10.3390/ijms23063346
3. Aprotosoaie AC, Costache AD, Costache II. Therapeutic Strategies and Chemoprevention of Atherosclerosis: What Do We Know and Where Do We Go? Pharmaceutics. 2022;14(4):722. doi:10.3390/pharmaceutics14040722
4. Kattoor AJ, Goel A, Mehta JL. LOX-1: Regulation, Signaling and Its Role in Atherosclerosis. Antioxidants. 2019;8(7):218. doi:10.3390/antiox8070218
5. Tuzcu EM, Kapadia SR, Tutar E, Ziada KM, Hobbs RE, McCarthy PM, Young JB, Nissen SE. High Prevalence of Coronary Atherosclerosis in Asymptomatic Teenagers and Young Adults: Evidence From Intravascular Ultrasound. Circulation. 2001;103(22):2705-2710. doi:10.1161/01.CIR.103.22.2705
6. Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ Res. 2016;118(4):535-546. doi:10.1161/CIRCRESAHA.115.307611
7. Shreya D, Zamora DI, Patel GS, Grossmann I, Rodriguez K, Soni M, Joshi PK, Patel SC, Sange I. Coronary Artery Calcium Score - A Reliable Indicator of Coronary Artery Disease? Cureus. Published online December 3, 2021. doi:10.7759/cureus.20149
8. Baeyens N, Bandyopadhyay C, Coon BG, Yun S, Schwartz MA. Endothelial fluid shear stress sensing in vascular health and disease. J Clin Invest. 2016;126(3):821-828. doi:10.1172/JCI83083
9. Kowara M, Cudnoch-Jedrzejewska A. Pathophysiology of Atherosclerotic Plaque Development-Contemporary Experience and New Directions in Research. Int J Mol Sci. 2021;22(7):3513. doi:10.3390/ijms22073513
10. Wang D, Yang Y, Lei Y, Tzvetkov NT, Liu X, Yeung AWK, Xu S, Atanasov AG. Targeting Foam Cell Formation in Atherosclerosis: Therapeutic Potential of Natural Products. Ma Q, ed. Pharmacol Rev. 2019;71(4):596-670. doi:10.1124/pr.118.017178
11. Pahwa R, Jialal I. Atherosclerosis. In: StatPearls. StatPearls Publishing; 2023. Accessed November 1, 2023. http://www.ncbi.nlm.nih.gov/books/NBK507799/
12. Bergheanu SC, Bodde MC, Jukema JW. Pathophysiology and treatment of atherosclerosis: Current view and future perspective on lipoprotein modification treatment. Neth Heart J. 2017;25(4):231-242. doi:10.1007/s12471-017-0959-2
13. Putra ED, Nazliniwaty N, Syafruddin S, Nerdy N. Component Analysis of White Ginger (Zingiber officinale Roscoe) Extract and Red Ginger (Zingiber officinale Rubra) Extract. Trop J Nat Prod Res. 2021;5(9):1634-1637. doi:10.26538/tjnpr/v5i9.17
14. Molecular Docking of Soybean (Glycine max) Seed and Ginger (Zingiber officinale) Rhizome Components as Anti-Diabetic Through Inhibition of Dipeptidyl Peptidase 4 (DPP-4) and Alpha-Glucosidase Enzymes. Trop J Nat Prod Res. 2021;5(10):1735-1742. doi:10.26538/tjnpr/v5i10.7
15. Ozkur M, Benlier N, Takan I, Vasileiou C, Georgakilas AG, Pavlopoulou A, Cetin Z, Saygili EI. Ginger for Healthy Ageing: A Systematic Review on Current Evidence of Its Antioxidant, Anti-Inflammatory, and Anticancer Properties. Gil G, ed. Oxid Med Cell Longev. 2022;2022:1-16. doi:10.1155/2022/4748447
16. FAO. FAOSTAT. 2023. Accessed November 13, 2023. https://www.fao.org/faostat/en/#data/QCL/visualize
17. Mao QQ, Xu XY, Cao SY, Gan RY, Corke H, Beta T, Li HB. Bioactive Compounds and Bioactivities of Ginger (Zingiber officinale Roscoe). Foods. 2019;8(6):185. doi:10.3390/foods8060185
18. Bischoff-Kont I, Fürst R. Benefits of Ginger and Its Constituent 6-Shogaol in Inhibiting Inflammatory Processes. Pharmaceuticals. 2021;14(6):571. doi:10.3390/ph14060571
19. Ma S qing, Guo Z, Liu F yuan, Hasan SG, Yang D, Tang N, An P, Wang M yu, Wu H ming, Yang Z, Fan D, Tang Q zhu. 6-Gingerol protects against cardiac remodeling by inhibiting the p38 mitogen-activated protein kinase pathway. Acta Pharmacol Sin. 2021;42(10):1575-1586. doi:10.1038/s41401-020-00587-z
20. Cerdá B, Marhuenda J, Arcusa R, Villaño D, Ballester P, Zafrilla P. Ginger in the Prevention of Cardiovascular Diseases. In: Shiomi N, Savitskaya A, eds. Current Topics in Functional Food. IntechOpen; 2022. doi:10.5772/intechopen.103970
21. Ballester P, Cerdá B, Arcusa R, Marhuenda J, Yamedjeu K, Zafrilla P. Effect of Ginger on Inflammatory Diseases. Molecules. 2022;27(21):7223. doi:10.3390/molecules27217223
22. Chen TC, Yen CK, Lu YC, Shi CS, Hsieh RZ, Chang SF, Chen CN. The antagonism of 6-shogaol in high-glucose-activated NLRP3 inflammasome and consequent calcification of human artery smooth muscle cells. Cell Biosci. 2020;10(1):5. doi:10.1186/s13578-019-0372-1
23. Suk S, Seo SG, Yu JG, Yang H, Jeong E, Jang YJ, Yaghmoor SS, Ahmed Y, Yousef JM, Abualnaja KO, Al-Malki AL, Kumosani TA, Lee CY, Lee HJ, Lee KW. A Bioactive Constituent of Ginger, 6-Shogaol, Prevents Adipogenesis and Stimulates Lipolysis in 3T3-L1 Adipocytes: Effects of 6-Shogaol on Adipogenesis and Lipolysis. J Food Biochem. 2016;40(1):84-90. doi:10.1111/jfbc.12191
24. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682-690. doi:10.1038/nchembio.118
25. Jiao W, Mi S, Sang Y, Jin Q, Chitrakar B, Wang X, Wang S. Integrated network pharmacology and cellular assay for the investigation of an anti-obesity effect of 6-shogaol. Food Chem. 2022;374:131755. doi:10.1016/j.foodchem.2021.131755
26. Luo L, Chen Y, Ma Q, Huang Y, Hong T, Shu K, Liu Z. Exploring the mechanism of an active ingredient of ginger, dihydrocapsaicin, on triple negative breast cancer based on network pharmacology and in vitro experiments. Oncol Lett. 2023;25(5):195. doi:10.3892/ol.2023.13781
27. Dai Y, Zhao Y, Nie K. The Antiemetic Mechanisms of Gingerols against Chemotherapy-Induced Nausea and Vomiting. Di Sotto A, ed. Evid Based Complement Alternat Med. 2022;2022:1-14. doi:10.1155/2022/1753430
29. Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Valencia A, ed. Bioinformatics. 2020;36(8):2628-2629. doi:10.1093/bioinformatics/btz931
30. Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545-D551. doi:10.1093/nar/gkaa970
31. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. PubChem 2023 update. Nucleic Acids Res. 2023;51(D1):D1373-D1380. doi:10.1093/nar/gkac956
32. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785-2791. doi:10.1002/jcc.21256
33. Ikechukwu K. Ijoma, Chinyere E. Okafor, Vincent I.E. Ajiwe. Computational Studies of 5-methoxypsolaren as Potential Deoxyhemoglobin S Polymerization Inhibitor. Trop J Nat Prod Res. 2024;8(10). doi:10.26538/tjnpr/v8i10.28
34. Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905-919. doi:10.1038/nprot.2016.051
35. Burley SK, Bhikadiya C, Bi C, Bittrich S, Chao H, Chen L, Craig PA, Crichlow GV, Dalenberg K, Duarte JM, Dutta S, Fayazi M, Feng Z, Flatt JW, Ganesan S, Ghosh S, Goodsell DS, Green RK, Guranovic V, et al. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023;51(D1):D488-D508. doi:10.1093/nar/gkac1077
36. BIOVIA. BIOVIA Discovery Studio. Published online 2021.
37. Seo T, Oelkers PM, Giattina MR, Worgall TS, Sturley SL, Deckelbaum RJ. Differential Modulation of ACAT1 and ACAT2 Transcription and Activity by Long Chain Free Fatty Acids in Cultured Cells. Biochemistry. 2001;40(15):4756-4762. doi:10.1021/bi0022947
38. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587-D592. doi:10.1093/nar/gkac963
39. Ivanova L, Karelson M. The Impact of Software Used and the Type of Target Protein on Molecular Docking Accuracy. Molecules. 2022;27(24):9041. doi:10.3390/molecules27249041
40. Kulandaisamy A, Lathi V, ViswaPoorani K, Yugandhar K, Gromiha MM. Important amino acid residues involved in folding and binding of protein–protein complexes. Int J Biol Macromol. 2017;94:438-444. doi:10.1016/j.ijbiomac.2016.10.04541.
41. Ferreira de Freitas R, Schapira M. A systematic analysis of atomic protein–ligand interactions in the PDB. MedChemComm. 2017;8(10):1970-1981. doi:10.1039/C7MD00381A
42. K. A. Thermodynamics of Ligand-Protein Interactions: Implications for Molecular Design. In: Moreno Pirajn JC, ed. Thermodynamics - Interaction Studies - Solids, Liquids and Gases. InTech; 2011. doi:10.5772/19447
43. Bohnacker T, Prota AE, Beaufils F, Burke JE, Melone A, Inglis AJ, Rageot D, Sele AM, Cmiljanovic V, Cmiljanovic N, Bargsten K, Aher A, Akhmanova A, Díaz JF, Fabbro D, Zvelebil M, Williams RL, Steinmetz MO, Wymann MP. Deconvolution of Buparlisib’s mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention. Nat Commun. 2017;8(1):14683. doi:10.1038/ncomms14683
44. Honda A, Kamata S, Akahane M, Machida Y, Uchii K, Shiiyama Y, Habu Y, Miyawaki S, Kaneko C, Oyama T, Ishii I. Functional and Structural Insights into Human PPARα/δ/γ Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate. Int J Mol Sci. 2022;23(9):4726. doi:10.3390/ijms23094726
45. Schnapp G, Neubauer H, Büttner FH, Handschuh S, Lingard I, Heilker R, Klinder K, Prestle J, Walter R, Wolff M, Zeeb M, Debaene F, Nar H, Fiegen D. A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state. Commun Chem. 2020;3(1):75. doi:10.1038/s42004-020-0321-2
46. Jang JY, Bae H, Lee YJ, Choi YI, Kim HJ, Park SB, Suh SW, Kim SW, Han BW. Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPARγ. Sci Rep. 2018;8(1):31. doi:10.1038/s41598-017-18274-1
47. Long T, Sun Y, Hassan A, Qi X, Li X. Structure of nevanimibe-bound tetrameric human ACAT1. Nature. 2020;581(7808):339-343. doi:10.1038/s41586-020-2295-8
48. Wang Z, Bao Z, Ding Y, Xu S, Du R, Yan J, Li L, Sun Z, Shao C, Gu W. Nε-carboxymethyl-lysine-induced PI3K/Akt signaling inhibition promotes foam cell apoptosis and atherosclerosis progression. Biomed Pharmacother. 2019;115:108880. doi:10.1016/j.biopha.2019.108880
49. Liu J, Xu P, Liu D, Wang R, Cui S, Zhang Q, Li Y, Yang W, Zhang D. TCM Regulates PI3K/Akt Signal Pathway to Intervene Atherosclerotic Cardiovascular Disease. Wang Y, ed. Evid Based Complement Alternat Med. 2021;2021:1-11. doi:10.1155/2021/4854755
50. Liu Z, Li J, Lin S, Wu Y, He D, Qu P. PI3K regulates the activation of NLRP3 inflammasome in atherosclerosis through part-dependent AKT signaling pathway. Exp Anim. 2021;70(4):488-497. doi:10.1538/expanim.21-0002
51. Zhao Y, Qian Y, Sun Z, Shen X, Cai Y, Li L, Wang Z. Role of PI3K in the Progression and Regression of Atherosclerosis. Front Pharmacol. 2021;12:632378. doi:10.3389/fphar.2021.632378
52. Wu B, Liu Y, Wu M, Meng Y, Lu M, Guo J, Zhou Y. Downregulation of microRNA‐135b promotes atherosclerotic plaque stabilization in atherosclerotic mice by upregulating erythropoietin receptor. IUBMB Life. 2020;72(2):198-213. doi:10.1002/iub.2155
53. Wang Y kai, Hong YJ, Yao Y hua, Huang XM, Liu XB, Zhang CY, Zhang L, Xu XL. 6-Shogaol Protects against Oxidized LDL-Induced Endothelial Injuries by Inhibiting Oxidized LDL-Evoked LOX-1 Signaling. Evid Based Complement Alternat Med. 2013;2013:1-13. doi:10.1155/2013/503521
54. Misawa K, Hashizume K, Yamamoto M, Minegishi Y, Hase T, Shimotoyodome A. Ginger extract prevents high-fat diet-induced obesity in mice via activation of the peroxisome proliferator-activated receptor δ pathway. J Nutr Biochem. 2015;26(10):1058-1067. doi:10.1016/j.jnutbio.2015.04.014
55. Han Q, Yuan Q, Xie G. 6-Shogaol attenuates LPS-induced inflammation in BV2 microglia cells by activating PPAR-γ. Oncotarget. 2017;8(26):42001-42006. doi:10.18632/oncotarget.16719
56. Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, Ji XL, Liu SQ. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int J Mol Sci. 2016;17(2):144. doi:10.3390/ijms17020144
57. Pantsar T, Poso A. Binding Affinity via Docking: Fact and Fiction. Molecules. 2018;23(8):1899. doi:10.3390/molecules23081899