Assessing Genetic Diversity in Squash Pumpkin (Cucurbita moschata) through Computational Analysis of Plastid Genes
Main Article Content
Abstract
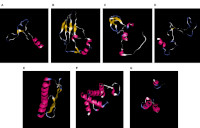
Grasping the genetic variation of economically significant crops, along with their Wild counterparts that serve as genetic assets, is essential for advancing the development of cultivars and strains capable of adapting to shifting climate conditions. Achieving high crop yields in agriculture is often challenging due to multiple influencing factors, such as cultivar quality, availability of nutrients and water, pathogen infection levels, natural disasters, and soil quality, all of which play a role in plant growth and development. This research evaluated the genetic diversity of 20 accessions of squash pumpkin (Cucurbita moschata) by analysing its’ plastid genes using bioinformatics mechanism. Twenty Cucurbita moschata accessions were sourced from the National Center for Biotechnology Information (NCBI) database. Phylogenetic association, Guanine-cytosine (GC) composition, secondary and three-dimensional protein structure, and domain structure were evaluated. The phylogenetic assessment identified 12 clades in total, with two main clades containing four accessions; XM_023105445.1, XM_023105444.1, XM_023075850.1, and XM_023075859.1 each showing 100% bootstrap support. Accession XM_021009681.1 showed the peak Guanine-cytosine composition, whereas XM_023076698.1 showed the least performance. The analysis of domain structure indicated that accession XM_02100968.1 contained the highest number of domains, while XM_023067309.1, XM_02105444.1, and XM_0230758951.1, each had only one domain, implying that they could have a more specific role. Variations in secondary and three-dimensional protein structures were observed across the accessions, suggesting potential structural differences that may affect protein robustness or their ability to interact with other molecules.
Downloads
Article Details
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
References
1. Wudil AH, Usman M, Rosak-Szyrocka J, Pilař L, Boye M. Reversing years for global food security: A review of the food security situation in Sub-Saharan Africa (SSA). Int J Environ Res Public Health. 2022; 19(22):14836. Doi: https://doi.org/10.3390/ijerph192214836
2. Ekerette EE, Etukudo OM, Efienokwu JN, Etta HE, Henry II, Ekpo PB, Edu EN, Agbor RB, Edem UL, Ikpeme EV. Evaluation of genetic variation in Oreochromis tilapia species from South-South Nigeria using mitochondrial DNA hypervariable region. Trop J Nat Prod Res. 2024; 8(9):8527-8536. Doi: https://doi.org/10.26538/tjnpr/v8i9.41.
3. Akerele D, Momoh S, Aromolaran AB, Oguntona CRB, Shittu AM. Food insecurity and coping strategies in South-West Nigeria. Food Sec. 2013; 5:407-414. Doi: https://doi.org/10.1007/s12571-013-0264-x.
4. Omorogiuwa O, Zivkovic J, Ademoh F. The role of agriculture in the economic development of Nigeria. Eur Sci J. 2014; 10:113-147.
5. Akinyele IO. Ensuring food and nutrition security in rural Nigeria: An assessment of the challenges, information needs, and analytical capacity (NSSP Working Paper No. 7). International Food Policy Research Institute (IFPRI); 2009.
6. Edem UL, Osuagwu AN, Udensi OU. Assessment of genetic diversity in Sphenostylis stenocarpa (Hoshst Ex. A Rich Harms) using morphological markers. Asian J Biol. 2023; 19(4):74-85.
7. Govindaraj M, Vetriventhan M, Srinivasan M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet Res Int. 2015; 2015:431487. Doi: https://doi.org/10.1155/2015/431487.
8. Rohman MS, Sulistomo HW, Kusumastuty I, Nugroho DA, Nugraheni NI, Lukitasari M, Alfata FH. In silico discovery of green tea and green coffee bioactive compounds against IGF-1R, PPAR-α, and TLR4 as a therapeutic candidate for metabolic disorder. Trop J Nat Prod Res. 2024; 8(3):6594–6603. Doi: https://doi.org/10.26538/tjnpr/v8i3.18. 9.
9. Kasemsap K. Bioinformatics: Applications and implications. In: Lytras M, Papadopoulou P, editors. Applying Big Data Analytics in Bioinformatics and Medicine. Idea Group Inc. Global; 2018. p. 26-47. Doi: https://doi.org/10.4018/978-1-5225-2607-0.ch002.
10. Oyebamiji AK, Akintelu AS, Mutiu OA, Adeosun IJ, Kaka MO, M T, Semire B. In-silico study on anti-cancer activity of selected alkaloids from Catharanthus roseus. Trop J Nat Prod Res. 2021; 5(7):1315–1322. Doi: https://doi.org/10.26538/tjnpr/v5i7.25.
11. Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011; 76(3-5):273-297. Doi: https://doi.org/10.1007/s11103-011-9762-4.
12. Choi H, Yi T, Ha SH. Diversity of Plastid Types and Their Interconversions. Front Plant Sci. 2021; 12:692024. Doi: https://doi.org/10.3389/fpls.2021.692024
13. Neuhaus HE, Emes MJ. Non-photosynthetic metabolism in plastids. Annu Rev Plant Biol. 2000; 51(1):111-140.
14. Edem UL, Osuagwu AN. Evaluation of secondary and tertiary protein structure variation in rbcL gene of selected legumes using a computational approach. J Proteomics Bioinformatics. 2023; 16(2):1000642.
15. Edem UL, Emeagi LI, Osuagwu AN, Edu NE, Ekerette EE, Ogbeche AA, Udensi OU. Assessment of genetic diversity analysis in African yam bean (Sphenostylis stenocarpa Hochst Ex.A.Rich Harms) using qualitative and quantitative attributes. J Adv Biol Biotechnol. 2024; 27(10):122-131. Doi: https://doi.org/10.9734/jabb/2024/v27i101436.
16. Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: Implications for crop plant evolution and areas of origin. Proc Natl Acad Sci U S A. 2002; 99(1):535-540. Doi: https://doi.org/10.1073/pnas.012577299.
17. Bowers JE, Tang H, Burke JM, Paterson AH. GC content of plant genes is linked to past gene duplications. PLoS One. 2022; 17(1):261-748. Doi: https://doi.org/10.1371/journal.pone.0261748.
18. Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000; 241(1):3-17. Doi: https://doi.org/10.1016/S0378-1119(99)00485-0.
19. Dufresne F, García-González F, Bell G. Genome size, environment, and genetic diversity in fish. Biol Lett. 2005; 1(4):411-414. Doi: https://doi.org/10.1098/rsbl.2005.0366.
20. Zhou Y, Zhou H. Prediction of protein secondary structure. Proteins Struct Funct Bioinform. 2004; 56(4):517-530. Doi: https://doi.org/10.1002/prot.2003.
21. Apic G, Gough J, Teichmann SA. Domain combinations in archeal, eubacterial, and eukaryotic proteomes. J Mol Biol. 2001; 310(2):311-325. https://doi.org/10.1006/jmbi.2001.4774.
22. Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018; 28(6):420-435. https://doi.org/10.1016/j.tcb.2018.02.004.